
Short description about ScikitLearn
Scikit-learn is a free machine learning library for Python. It features various algorithms like support vector machine, random forests, and k-neighbors, and it also supports Python numerical and scientific libraries like NumPy and SciPy.
Moving to Questions and Answers
1. What is Data Leakage and how do you avoid it in Scikit-Learn?
Answer:

2. What are the basic objects that a ML pipeline should contain?
Answer:

3. What is the difference between the Recursive Feature Elimination (RFE) function and SelectFromModel in Scikit-Learn?
Answer: They effectively try to achieve the same result but the methodology used by each technique varies a little:
- RFE removes least significant features over iterations. So basically it first removes a few features which are not important and then fits and removes again and fits. It repeats this iteration until it reaches a suitable number of features.
- SelectFromModel is a little less robust as it just removes less important features based on a threshold given as a parameter. There is no iteration involved.
4. What’s the difference between StandardScaler and Normalizer and when would you use each one?
Answer:
- With Normalize each sample (i.e. each row of the data matrix) with at least one non-zero component is rescaled independently of other samples so that its norm (l1 or l2) equals one.
- On another hand, StandardScaler standardizes features by removing the mean and scaling to unit variance.
- In other words Normalizer acts row-wise and StandardScaler column-wise. Normalizer does not
- remove the mean and scale by deviation but scales the whole row to unit norm.
5. What’s the problem if you call the fit() method multiple times with different X and y data? How can you overcome this issue?
Answer: Given an initial model, as soon you call model.fit(features_train, label_train) the model starts training using the features and labels that you have passed. If you will again call model.fit(features_train2, label_train2) it will start training again using passed data and will remove the previous results. The model will reset the following inside model:
- Weights
- Fitted Coefficients
- Bias, etc.
To overcome this issue we can use partial_fit() method as well if we want our previous calculated stuff to stay and additionally train using the next data.
6. How are feature_importances_ in RandomForestClassifier determined in Scikit-Learn?
Answer: The usual way to compute the feature importance values of a single tree is as follows:
You initialize an array feature_importances of all zeros with size n_features.
You traverse the tree: for each internal node that splits on feature i you compute the error reduction of that node multiplied by the number of samples that were routed to the node and add this quantity to feature_importances[i].
The error reduction depends on the impurity criterion that you use (in Scikit-learn the criterion available are Gini and entropy). It’s the impurity of the set of examples that gets routed to the internal node minus the sum of the impurities of the two partitions created by the split.
Its important that these values are relative to a specific dataset (both error reduction and the number of samples are dataset specific) thus these values cannot be compared between different datasets.
7. How can you obtain the principal components and the eigenvalues from Scikit-Learn PCA?
Answer:

8. How do you scale data that has many outliers in Scikit-Learn?
Answer:
If the data contains many outliers, scaling using the mean and variance of the data is likely to not work very well. In these cases, we can use RobustScaler as a drop-in replacement instead.
RobustScaler removes the median and scales the data according to the quantile range (defaults to IQR: Interquartile Range). The IQR is the range between the 1st quartile (25th quantile) and the 3rd quartile (75th quantile).
Centering and scaling happen independently on each feature by computing the relevant statistics on the samples in the training set. Median and interquartile ranges are then stored to be used on later data using the transform method.
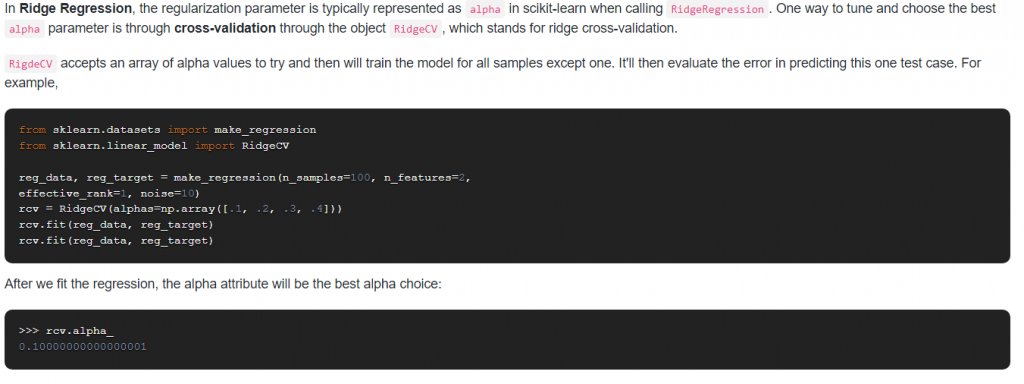
9. How do you optimize the Ridge Regression parameter?
Answer:
10. How would you encode a large Pandas dataframe using Scikit-Learn?
Answer:

11. Is max_depth in Scikit-learn the equivalent of pruning in decision trees? If not, how a decision tree is pruned using scikit?
Answer:
- Though they have similar goals (i.e. placing some restrictions to the model so that it doesn’t grow very complex and overfit), max_depth isn’t equivalent to pruning.
- The way pruning usually works is that go back through the tree and replace branches that do not help with leaf nodes.
- However, scikit-learn does pruning based on minimal cost-complexity pruning: the subtree with the largest cost complexity that is smaller than the ccp_alpha parameter will be chosen.
12. Suppose you have multiple CPU cores available, how can you use them to reduce the computational cost?
Answer:
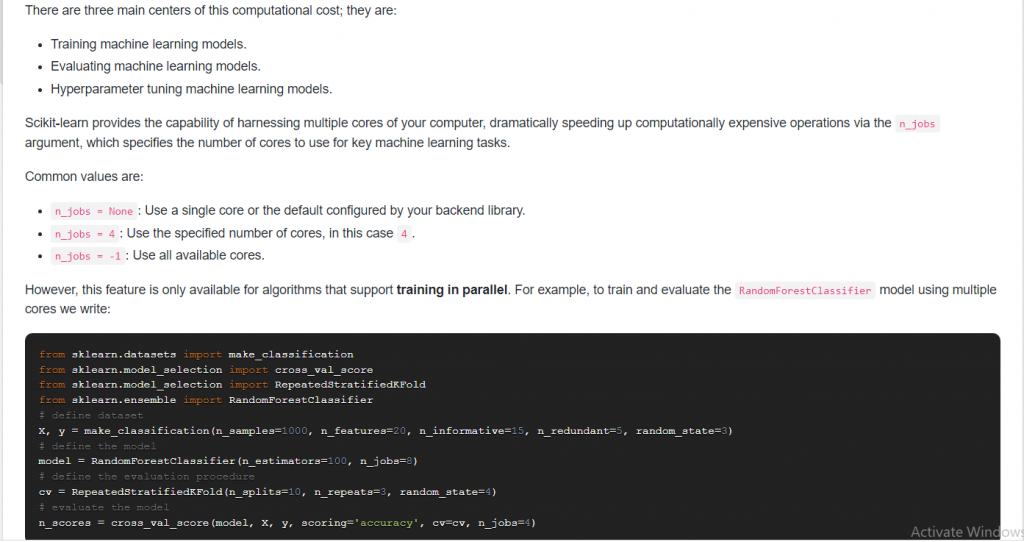
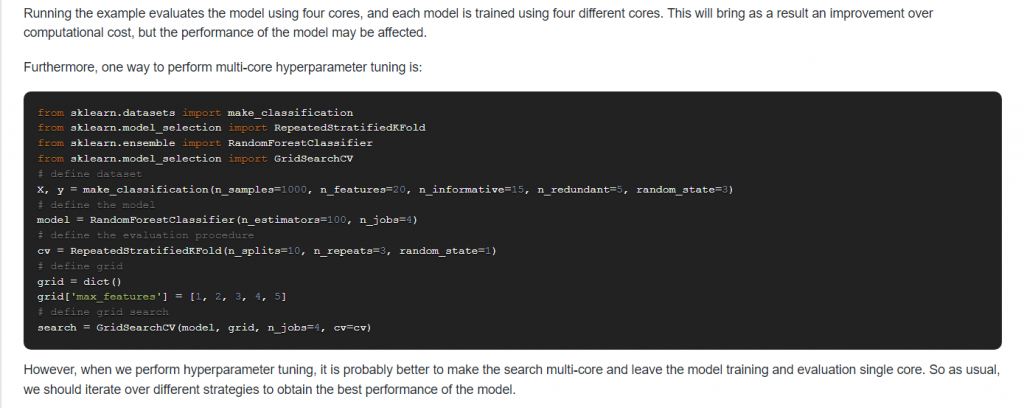
13. What is the difference between cross_validate and cross_val_score in Scikit-Learn?
Answer:
- cross_val_score calculates the score for each CV split and returns an array of scores of the estimator for each run of the cross validation.
- cross_validate it allows specifying multiple metrics for evaluation, i.e. calculate one or more scores and timings for each CV split and it returns dict containing training scores, fit-times and score-times in addition to the test score.
14. What’s the difference between fit(), transform() and fit_transform()? Why do we need these separate methods?
Answer:
- Every Scikit-Learn’s transform’s fit() calculates the model parameters (e.g. μ and σ in case of StandardScaler) and saves them as an internal object’s state.
- Afterwards, you can call its transform() method to apply the transformation to any particular set of examples.
- fit_transform() joins these two steps and is used for the initial fitting of parameters on the training set x, while also returning the transformed x’. Internally, the transformer object just calls first fit() and then transform() on the same data.
In practice, we need to have separate training and testing dataset and that is where having a separate fit() and transform() method helps. We apply fit() on the training dataset and use the transform() method on both -the training dataset and the test dataset-. Thus the training, as well as the test dataset, are then transformed (scaled) using the model parameters that were learned on applying the fit() method to the training dataset.
So for the x training set, we do fit_transform because we need to compute model parameters, and then use it to autoscale the data. For the x test set, well, we already have the model parameters, so we only do the transform part.
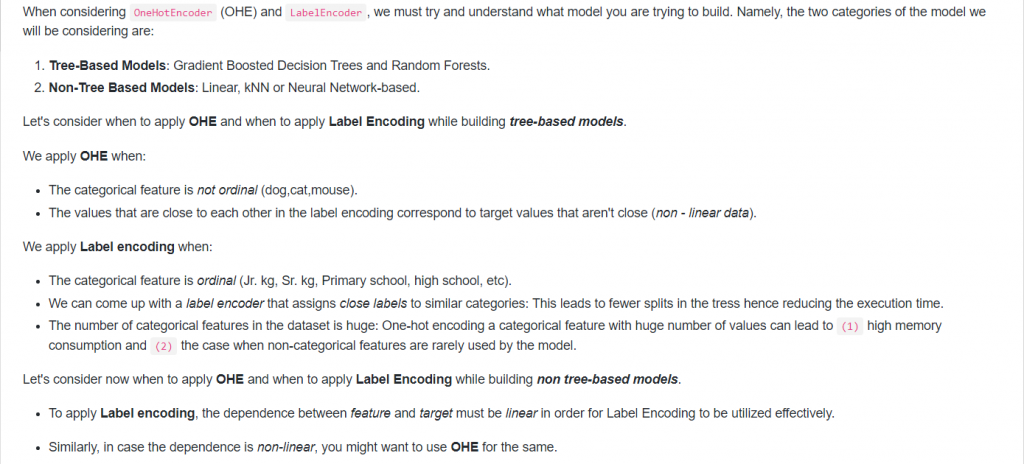
15. When to use OneHotEncoder vs LabelEncoder in Scikit-Learn?
Answer:
16. While using KNeighborsClassifier, when would you set weights=”distance”?
Answer:
- weights = ‘distance’ is in contrast to the default which is weights = ‘uniform’.
- When weights are uniform, a simple majority vote of the nearest neighbors are used to assign cluster membership.
- When weights are distance weighted, the voting is proportional to the distance value. Nearby points will have a greater influence than more distance points (even if the counts of different groups are similar). Therefore distance weighting is very useful when we have sparse data.
17. Would you use PCA on large datasets or there is a better alternative?
Answer: The PCA object is very useful but has certain limitations for large datasets. The biggest limitation is that PCA only supports batch processing, which means all of the data to be processed must fit in the main memory.
A better alternative to using with the large dataset is IncrementalPCA, this object uses a different form of processing and allows for partial computations which almost exactly match the results of PCA while processing the data in a minibatch fashion.
IncrementalPCA has the parameter batch_size to specify the number of samples to use for each batch and only stores estimates of component and noise variances, in order to update explained_variance_ratio_ incrementally. The memory usage depends on the number of samples per batch, rather than the number of samples to be processed in the dataset.
Therefore depending on the size of the input data, this algorithm can be much more memory efficient than a PCA and allows sparse input.
18. Are there any advantages of XGBoost over GradientBoostingClassifier?
Answer: XGBoost and sklearn’s GradientBoost are fundamentally the same as they are both gradient boosting implementations. However, there are very significant differences under the hood in a practical sense. XGBoost is a lot faster and this comes from the fact that XGBoost uses an approximation on the split points.
For example, if you have a continuous feature with 10000 possible splits, XGBoost considers only the best 300 splits by default. This behavior is controlled by the sketch_eps parameter whose default value is 0.03. We can also try lowering it and checking the difference it makes. In addition to that, XGBoost can be parallelized.
In sci-kit-learn documentation there is no mention of a sketch_eps-like feature, so we could infer that is not available. On other hand, GradientBoostingClassifier does not have a n_jobs interface parameter available, therefore parallel processing is not possible internally with sklearn’s implementation of the algorithm.
Given all the above features, XGBoost has the advantage of being memory-efficient and its performance is superior to sklearn’s GradientBoosting.
19. Can you use SVM with a custom kernel in Scikit-Learn?
Answer: Yes, we can use our own defined kernels by passing a function to the kernel parameter.
Our custom kernel must take as arguments two matrices of shape (n_samples_1, n_features), (n_samples_2, n_features) and return a kernel matrix of shape (n_samples_1, n_samples_2).
For example,
import numpy as np
from sklearn import svm
def my_kernel(X, Y):
… return np.dot(X, Y.T)
…
clf = svm.SVC(kernel=my_kernel)
20. How does sklearn KNeighborsClassifier compute class probabilites when setting weights=’uniform’?
Answer:
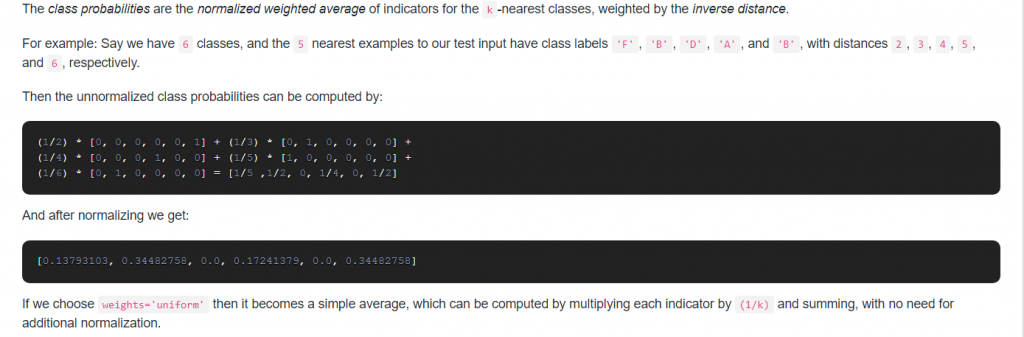
21. When you would use StratifiedKFold instead of KFold?
Answer:
- KFold is a cross-validator that divides the dataset into k folds. If shuffle is set to False, consecutive folds will be the shifted version of the previous fold. If shuffle is set to True, then the splitting will be random.
- StratifiedKFold takes the cross-validation one step further. The class distribution in the dataset is preserved in the training and test splits, i.e. each fold of the dataset has the same proportion of observations with a given label.
Therefore, we should prefer StratifiedKFold over KFold when dealing with classifications tasks with imbalanced class distributions.
22. How to predict time series in scikit-learn?
Answer: Time Series is a collection of data points collected at constant time intervals. Time-series prediction is based on the theory that the current values more or less depends on the past ones. A time series has two basic components: Mean and Variance. Ideally, you would like to control this component, for the variability, you can simply apply a logarithm transformation on the data, and for the trend, you can differentiate it.
In the case of prediction of time series data, RNN or LSTM algorithm (Deep Learning) has been widely utilized, but scikit does not provide the built-in algorithm of it. So, you might be better off studying Tensorflow or Pytorch framework which are common tools to enable you to build the RNN or LSTM model.
23. Is any custom distance function using scikit-learn K-Means Clustering?
Answer: Unfortunately no: by definition, the k-means clustering algorithm relies on the euclidean distance from the mean of each cluster. It has no metric parameter and it is not trivial to extend k-means to other distances. You could use a different metric, so even though you are still calculating the mean you could use something like the Mahalanobis distance.
24. What does the “fit()” method in scikit-learn do?
Answer: Fitting your model (using fit() method) to the training data is essentially the training part of the modeling process. The fit() method finds the coefficients for the equation specified via the algorithm being used. During the process, this method modifies the object and returns a reference to the object. After it is trained, the model can be used to make predictions, usually with a .predict() method call.
25. Is it possible in scikit-learn to split into three sets directly?
Answers:

I’m a DevOps/SRE/DevSecOps/Cloud Expert passionate about sharing knowledge and experiences. I am working at Cotocus. I blog tech insights at DevOps School, travel stories at Holiday Landmark, stock market tips at Stocks Mantra, health and fitness guidance at My Medic Plus, product reviews at I reviewed , and SEO strategies at Wizbrand.
Please find my social handles as below;
Rajesh Kumar Personal Website
Rajesh Kumar at YOUTUBE
Rajesh Kumar at INSTAGRAM
Rajesh Kumar at X
Rajesh Kumar at FACEBOOK
Rajesh Kumar at LINKEDIN
Rajesh Kumar at PINTEREST
Rajesh Kumar at QUORA
Rajesh Kumar at WIZBRAND

 Starting: 1st of Every Month
Starting: 1st of Every Month  +91 8409492687
+91 8409492687  Contact@DevOpsSchool.com
Contact@DevOpsSchool.com
